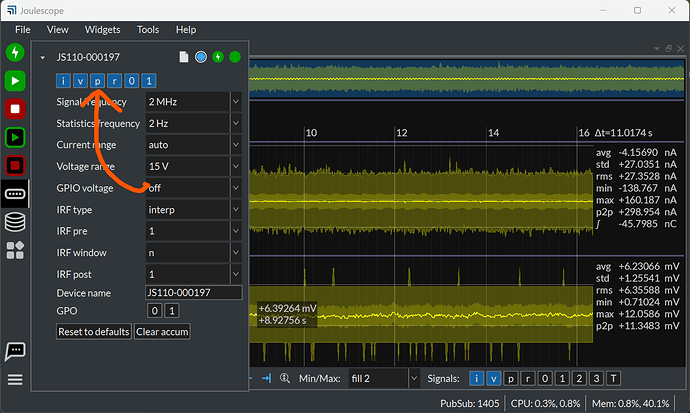
I captured i,v,p,0,1 data using pyjoulescope_driver.
① When reading out the captured jls file, current data has “nan” in 1st/2nd data for a while at the beginning (and 3rd/4th data are quite big).
② Also, data length is different.
I set --duration 5 --frequency 1000000, then I got;
- gpi[0], gpi[1] : 5000000 (expected)
- power/voltage : 5003152
- current : 5019396.
I wonder why current data is strange at the beginning and different length in signals.
I appreciate your advice.
I also captured data using UI for comparison.
There is no “nan” or big value in current, although length is different between voltage/current and gpi[0]/gpi[1]
③ (I couldn’t select power data in UI, guessing it’s limitation in current version)
Environment:
- Joulescope JS110
- python 3.10.9
- python modules: joulescope 1.1.3, pyjoulescope_driver 1.3.3, pyjls 0.5.3
- Joulescope UI 1.0.9 (alpha)
- command: python -m pyjoulescope_driver record --set “s/i/lsb_src=gpi0” --set “s/v/lsb_src=gpi1” --set “s/extio/voltage=1.8V” --duration 5 --frequency 1000000 --signals i,v,p,0,1 test.jls
Here is a test script to read out jls file.
from pyjls import Reader
jls_path = "test.jls"
signal_list = ['power', 'voltage', 'current', 'gpi[0]', 'gpi[1]']
r = Reader(jls_path)
for signal in signal_list:
signals = [s for s in r.signals.values() if s.name == signal]
if len(signals) == 0:
print(f"no {signal} data.")
continue
data = r.fsr_statistics(signals[0].signal_id, 0, 1, signals[0].length)
print(f"signal={signal}, length={signals[0].length}")
print(f"data[0:5]=\n{data[0:5]}")
Result
[jls file from pyjoulescope_driver]
signal=power, length=5003152
data[0:5]=
[[0.00247067 0. 0.00247067 0.00247067]
[0.00247074 0. 0.00247074 0.00247074]
[0.00247058 0. 0.00247058 0.00247058]
[0.00247082 0. 0.00247082 0.00247082]
[0.00247203 0. 0.00247203 0.00247203]]
signal=voltage, length=5003152
data[0:5]=
[[1.82927263 0. 1.82927263 1.82927263]
[1.82927084 0. 1.82927084 1.82927084]
[1.82927585 0. 1.82927585 1.82927585]
[1.82926738 0. 1.82926738 1.82926738]
[1.82927716 0. 1.82927716 1.82927716]]
signal=current, length=5019396
data[0:5]=
[[ nan nan 1.79769313e+308 -1.79769313e+308]
[ nan nan 1.79769313e+308 -1.79769313e+308]
[ nan nan 1.79769313e+308 -1.79769313e+308]
[ nan nan 1.79769313e+308 -1.79769313e+308]
[ nan nan 1.79769313e+308 -1.79769313e+308]]
signal=gpi[0], length=5000000
data[0:5]=
[[1. 0. 1. 1.]
[1. 0. 1. 1.]
[1. 0. 1. 1.]
[1. 0. 1. 1.]
[1. 0. 1. 1.]]
signal=gpi[1], length=5000000
data[0:5]=
[[0. 0. 0. 0.]
[0. 0. 0. 0.]
[0. 0. 0. 0.]
[0. 0. 0. 0.]
[0. 0. 0. 0.]]
[jls file from JouleScope UI(1.0.9)]
no power data.
signal=voltage, length=13839888
data[0:5]=
[[1.82472885 0. 1.82472885 1.82472885]
[1.82700062 0. 1.82700062 1.82700062]
[1.82700062 0. 1.82700062 1.82700062]
[1.82700062 0. 1.82700062 1.82700062]
[1.82927239 0. 1.82927239 1.82927239]]
signal=current, length=13839888
data[0:5]=
[[0.00169455 0. 0.00169455 0.00169455]
[0.0018052 0. 0.0018052 0.0018052 ]
[0.00190987 0. 0.00190987 0.00190987]
[0.00198763 0. 0.00198763 0.00198763]
[0.00204744 0. 0.00204744 0.00204744]]
signal=gpi[0], length=13800000
data[0:5]=
[[1. 0. 1. 1.]
[1. 0. 1. 1.]
[1. 0. 1. 1.]
[1. 0. 1. 1.]
[1. 0. 1. 1.]]
signal=gpi[1], length=13800000
data[0:5]=
[[0. 0. 0. 0.]
[0. 0. 0. 0.]
[0. 0. 0. 0.]
[0. 0. 0. 0.]
[0. 0. 0. 0.]]